Table Of Content

Primer Tm values were compared between STITCHER 2.0 and primer design software, Primer310,11. Batch Primer318 was used to generate primer Tm values for 544 primers and compared with the values calculated by STITCHER 2.0. Correlation of the data sets revealed an R-squared value of 0.98. It is not always possible to generate primers specific to a particular splice variant mRNA when the difference in exons is not sufficient to distinguish one from the rest.
Designing reverse primer
The 'Couple' strategy is one step further of the '2-Round' approach, that further divides the assembly into sub-pools of primer pairs. It is rare when neighboring primers have mispriming sites other than the designed region. Cerevisiae was used as a template to amplify the GAL3 gene, which encodes a protein involved in galactose metabolism. The goal for this experiment was to determine the optimal Mg2+ concentration for this set of reagents. No MgCl2 was present in the original PCR buffer and had to be supplemented at the concentrations indicated with a range tested from 0.0 mM to 5.0 mM.
For Questions Related to NEB Products and Offers

In addition to checking for amplicons between the forward and the reverse primers, Primer-BLAST also checks amplicons arising from either primer alone. For example, the forward primer could also act as a reverse primer if it happens to match some regions on the minus strand of the template. It is therefore desirable to combine various elements of primer design requirements into one process such that users can simply input the template and obtain the desired target-specific primers.
Multiplex real-time RT-PCR method for the diagnosis of SARS-CoV-2 by targeting viral N, RdRP and human RP genes ... - Nature.com
Multiplex real-time RT-PCR method for the diagnosis of SARS-CoV-2 by targeting viral N, RdRP and human RP genes ....
Posted: Fri, 18 Feb 2022 08:00:00 GMT [source]
INTRODUCTION TO REAL TIME POLYMERASE CHAIN REACTION (RT...
The 5′- and 3′-ends of the primers play different roles; although the 5′-end contributes marginally, matching the 3′ end is critical for PCR efficiency amplification. Multiple pairs of primers in the same amplification system can also increase the probability of primer dimer formation, which can be predicted by related software such as Oligo7 57. The bases of primers should be randomly distributed, and it is suggested that there should be no more than four complementary bases between the forward and reverse primers.
HOW TO DESIGN PRIMERS FOR PCR AND QUANTITATIVE REAL TIME PCR (QPCR)
Thus, cytosines that were methylated in the original DNA will be the only cytosines left in the DNA sequence after amplification. Based on this principle it is possible, by sequencing the bisulfite PCR product, to identify the methylation status of the cytosines in the original DNA sample. While this method is useful to learn the methylation status of DNA, the conversion of unmethylated cytosines to uracil causes the two DNA strands to lose complementarity and strongly decreases the complexity of the DNA sequence. This makes the design of primers specific for the DNA region of interest more challenging. These features make STITCHER 2.0 novel and provides an intuitive interface to design robust primers, and offers comprehensive resources during experimental design and troubleshooting. The first file returns a compositional analysis of the input sequence.
HOW TO DESIGN PRIMERS FOR YOUR PCR EXPERIMENT
The addition of a GC clamp to either the forward or reverse primer can be specified (by default this is set to ‘No’). During primer design STITCHER 2.0 will move along in increments defined by the user within the specified 5′ and 3′ search areas. The default increment is 2 bp, however, the forward and reverse increments can be modified independently to facilitate specific applications.
Exon/Intron Selection
Note that this option cannot be used in association with the "Exon/intron selection" options above. Designing oligonucleotides and making sure that you have the right parameters for your oligo is an important step in securing results, especially in PCR Primer Design. In order to achieve successful DNA amplification, it’s important to start off with the right primer. To reverse complement, copy the sequence (ctrl+c or cmd+c) and past the sequence (ctrl+v or cmd+v) in new window (Fig 4.1). There is a button in the header part of the APE to reverse complement the sequence. Select the primer sequence (if it is not selected already) and click on the button (Fig 4.2).
Next, tables are returned of the designed forward primers, reverse primers, and compatible primers as well as various metrics describing the forward and reverse primers designed by STITCHER 2.0. A graphic of the primer sequence map is also generated, and finally, an intron-exon map of the input sequence is generated which can be used to draw primers in specific locations. Polymerase Chain Reaction (PCR) is a technique that has various applications in research, medical, and forensic field. It is also a sensitive test for disease diagnosis and genotyping. The basic ingredients of a reaction system include a DNA template, a buffer solution, deoxyribonucleoside triphosphate (dNTPs), Taq polymerase, and a pair of primers (the forward one and the reverse one).
The following table gives the comparison of primers designed for various cloning methods. Primers are short stretches of DNA that target unique sequences of a DNA molecule and help identify a unique part of a genome or a gene to be copied further. Primers can be synthesized and used for your PCR (polymerase chain reaction) and other molecular biology experiment such as in transformation experiment, gene cloning experiment, gene amplification experiment etc. PCR experiment is a DNA amplification technique in which millions of DNA copies are made using a single DNA segment. Common problems that arise with primers and 3-step PCR amplification of target DNA.
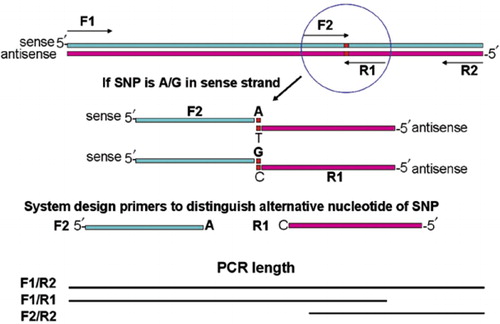
Also, the most common adjustments that are required for optimizing a PCR experiment are to change the Mg2+ concentration and to correct the annealing temperatures. However, if these changes do not minimize or abrogate aberrant results, titration of additives and /or changing the cycling condition protocols as described in Tables 2-6 may alleviate the problem. Primer design is a critical first step in the design of a quality real time PCR experiment. There are many primer design elements to consider that can help minimize troubleshooting down the line, including primer specificity, primer size and amplicon length. Read on to learn how to create specific and effective primers for several types of PCR assays including real time quantitative PCR (qPCR), reverse transcription quantitative PCR (RT-qPCR), bisulfite PCR, and methylation specific PCR (MSP). Illustration depicting the design of reverse primer and its involvement in PCR.
The example primers correspond to those underlined in Additional files 1 and 2. Enabling this option will make it much easier to find gene-specific primers since there is no need to distinguish between splice variants. This option requires you to enter a refseq mRNA accession or gi or fasta sequence as PCR template input because other type of input may not allow the program to properly interpret the result. Primer-BLAST also offers the capability to design primers based on exon/intron structure so that PCR amplification can better be targeted to mRNA. Users can specify whether a primer should span an exon/exon junction with an adjustable number of bases on each side of the junction and whether the primer pair should span an intron along with an option to specify intron size.

No comments:
Post a Comment